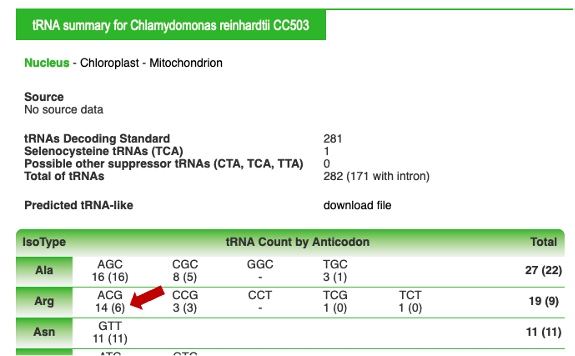
Example 1 : Looking for all nuclear-encoded Arabidopsis thaliana tRNAArg
Possibility 1: From the Home page click on "tRNA"

Select "Vitis vinifera"

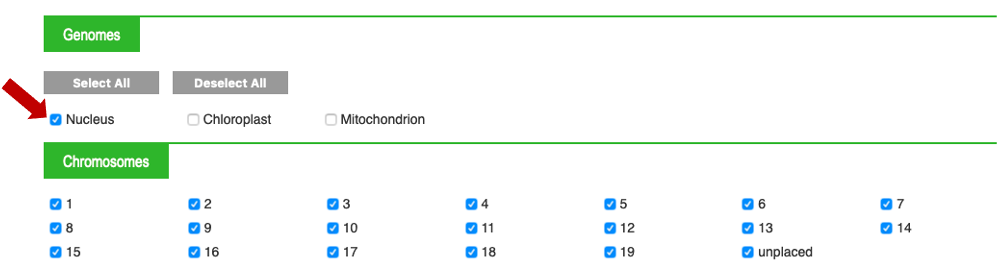
Select "Nucleus" (all chromosomes are selected by default, if you wish only one, deselect the ones you do not want)
Select "Arg": (by default, all anticodons are selected, if you wish to restrict your choice, deselect the anticodons you are not interested in)

Go to the bottom of the page, click on "search in database"
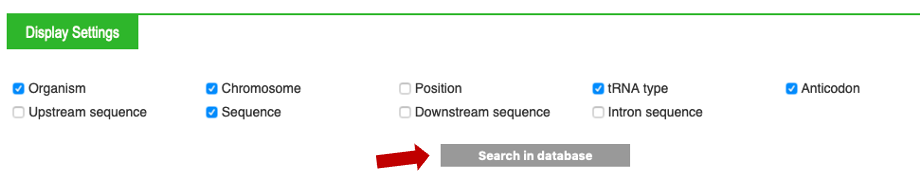
Remark : you have several options for display settings !
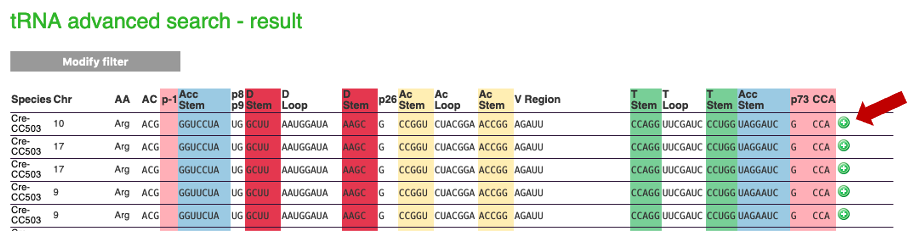
Results are displayed (you may have several pages), and can be downloaded (xls or fasta format)
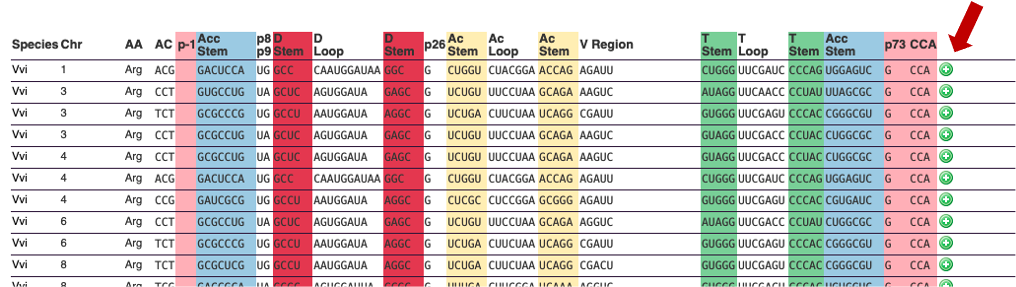
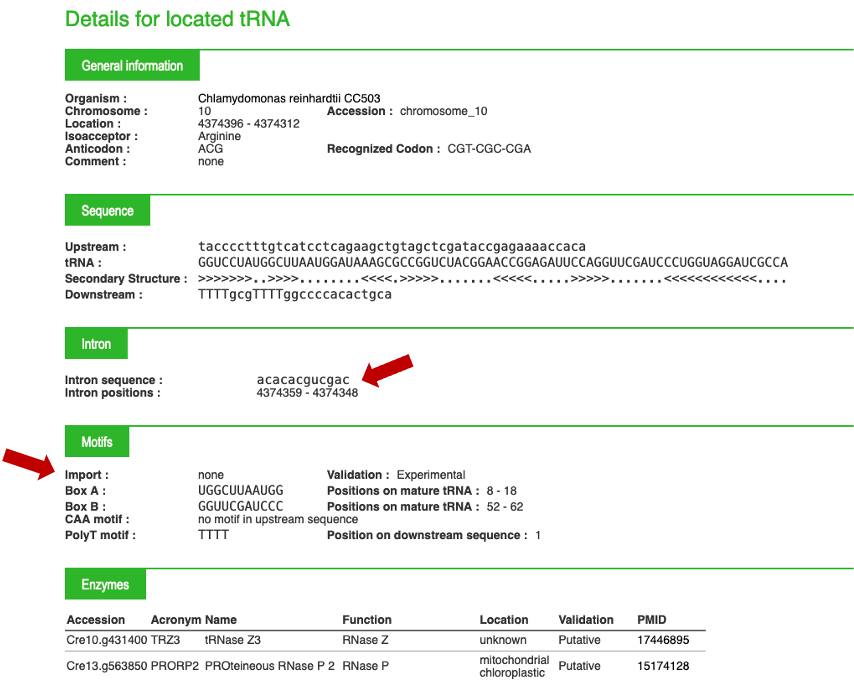
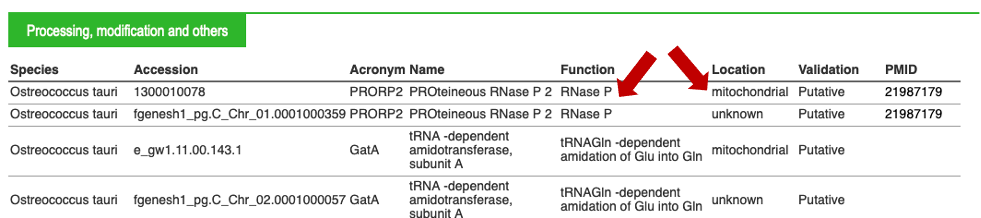
For each tRNA, you can click on the "+" button on the right of the screen. This "+" will give you access to important biological information, an example is given below:
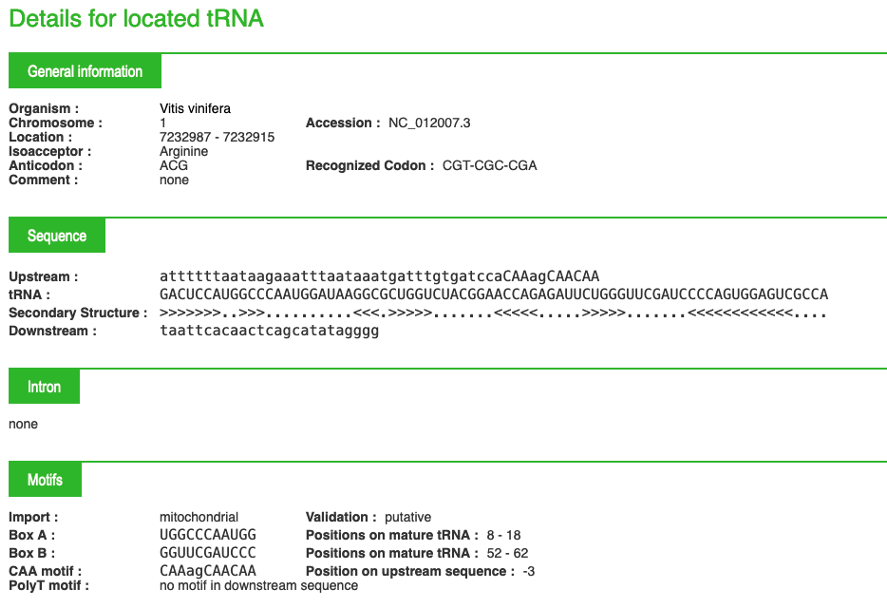
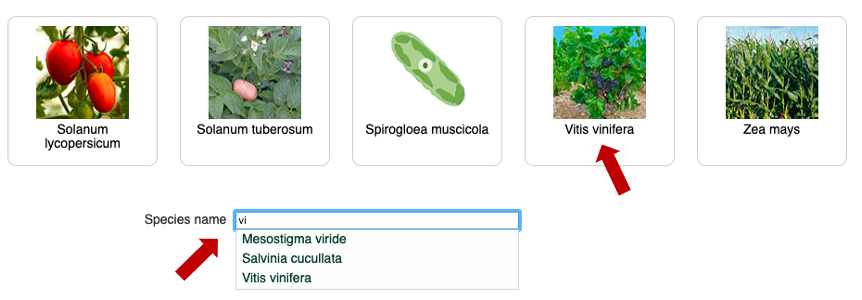
Possibility 2: From the Home page click on "Species"

Then, click on "Vitis vinifera" picture or begin to type the species name in filter input then select it in the drop down list
On the "Arg" line, click on the number on the right (total column) to obtain the complete list of A. thaliana tRNAArg genes
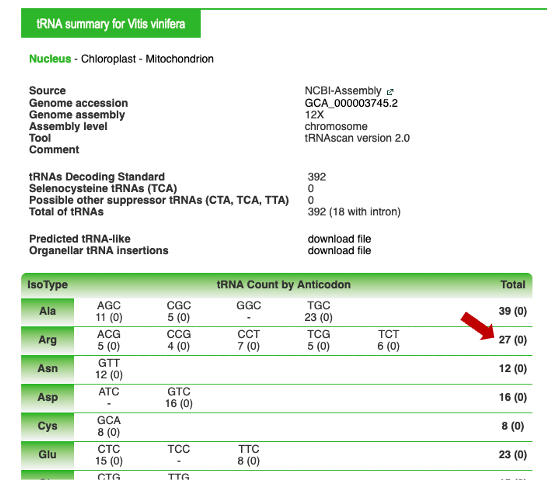

By clicking on the "+" button, additional information can be provided, as described in example 1. Clicking on "arath", on the left, will allow you to go back to the summary for the species
 PlantRNAInstitut de biologie moléculaire des plantes
PlantRNAInstitut de biologie moléculaire des plantes